Casilio
Navigation: [Module Registry][sgRNA-PBS][The paper]
Subprojects: [Casilio-ME]
CRISPRzymes
RNA-guided nuclease CRISPR/Cas9 is widely used in genome editing [1-5]. Nuclease-deficient mutant dCas9 can be repurposed as a versatile RNA-programmable DNA-binding domain that does not generate breaks on DNA [6]. CRISPR/dCas9 has been used to bind target DNA and block transcription, activate or repress genes, tether epigenetic factor such as histone acetyltransferase to modify epigenetic status [7-11] and visualize genomic loci in live cells by tethering fluorescent proteins to defined loci [12]. These CRISPR/dCas9-based enzymes (CRISPRzymes) enable researchers to perform functional studies in situ and in vivo without permanently changing genetic sequences.
The problems and previous solutions
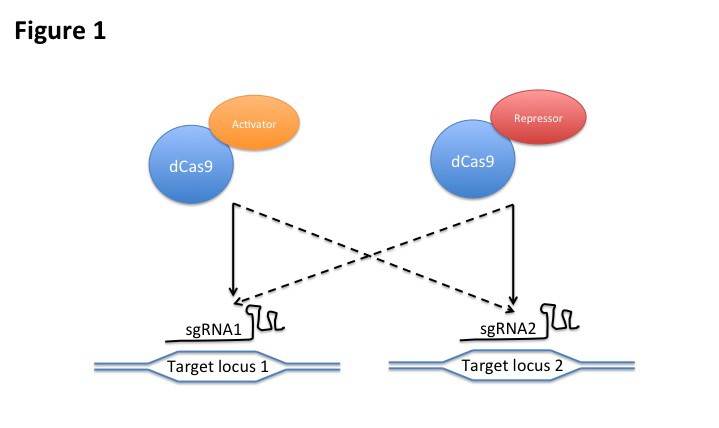
It is often useful to deliver distinct functions to separate genomic loci, such as, activation of a set of genes and repression of another set of genes. Although dCas9 can tether different effectors, several dCas9 fusions (e.g., dCas9-VP64 activator and dCas9-KRAB repressor) cannot be co-introduced with sgRNAs to regulate two set of genes differently since both fusions will bind to both set of genes directed by both sets of sgRNAs (Fig 1). Several solutions have been developed to circumvent such undesired crosstalks. Orthogonal CRISPR/dCas9 derived from different bacterial species that utilize sgRNAs with different repeat sequences and recognize different PAM sequences can be fused to distinct effectors and targeted to distinct sets of genomic targets [13]. Another strategy involves the use of a modified sgRNA extended with structural aptamers (e.g., MS2 and PP7) that can recruit corresponding RNA binding domain fused to effector proteins [14-17]. However, limited number of natural aptamers have been identified and characterized. On the other hand, more than one molecule of effector proteins is often desired to achieve sufficient biological activity. While structured RNA aptamers can be multiplied on the modified RNA, it have been shown that limited number of these structured elements can be inserted on the sgRNAs before being inhibitory to functions. Another solution is to use Suntag, a technology relying on the fusion of multiple copies of epitope tags to the dCas9 protein to recruit antibody-effector fusions [18]. However, limited number of antibody/epitope pairs suitable for such engineering have been reported. Suntag also does not enable multiplexing since it is directly fused to the dCas9 protein.
Our solution to both the multiplexing and multimerization problems – Casilio
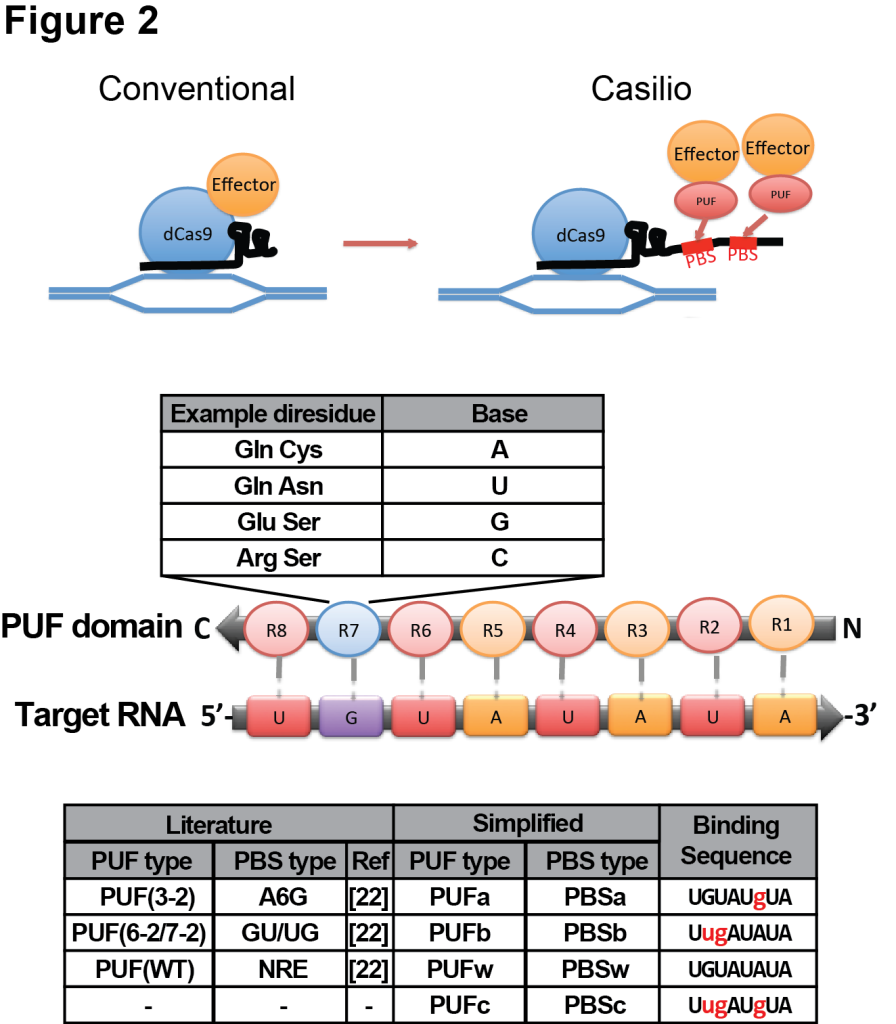
We have constructed an RNA-aptamer based approach called Casilio to improve both multiplexing and multimerization capabilities to CRISPRzyme toolkits [19]. Casilio is a hybrid system combining CRISPR/dCas9 and Pumilio RNA-binding domain (Fig 2). Pumilio/PUF RNA binding domain contains 8 sequence repeats each recognizing one RNA base [20-23]. Specific diresidues in each repeat can be changed to enable recognition of each of the 4 RNA bases. As opposed to directly fusing effector domains to dCas9 in conventional CRISPR/dCas9-based enzymes, we relocated the tethering of effector proteins to a modified sgRNA (sgRNA-PBS) with different numbers of 8-mer pumilio binding sites (PBS) which can recruit effector domains fused to cognate PUF domains.
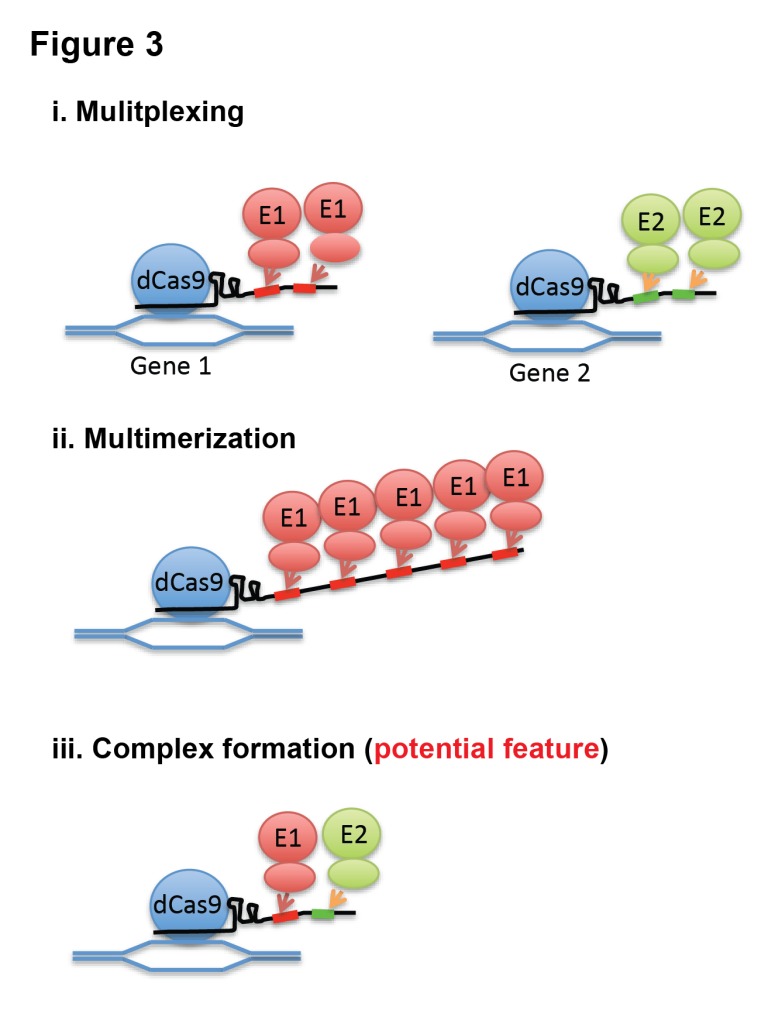
With this feature, we demonstrated that Casilio can multiplex and multimerize effectors and potentially enable assembly of stoichiometrically defined protein complexes (Fig 3). We showed that Casilio modules are orthogonal, allowing simultaneous and independent activation and repression of different genes. We showed that Casilio enables multicolor imaging of telomeres and centromeres. We also showed epigenetic editing of enhancers by tethering histone acetyltransferase domain of CBP by Casilio system. We demonstrated that, by inserting multiple binding sites and thus recruiting multiple molecules of effectors or fluorescent proteins, we enhanced magnitude of gene regulation, epigenetic editing, as well as signal-to-noise ratio of chromosome labeling. For more details, please read our Open Access paper.
The components of Casilio (using Sp-dCas9)
Sp-dCas9 – nuclease-deficient S. pyogenes Cas9 with D10A and H840A mutations
pAC1445 – pmax-dCas9 – Transient expression vector for dCas9
Other dCas9-expressing vectors may be available from Addgene
sgRNA-PBS – A single-guide RNA (sgRNA) appended with multiple copies of Pumilio binding sites (PBS).
List of vectors expressing sgRNA-PBS
Casilio modules – A fusion protein consisting of effector domain(s) fused with a PUF domain programmed to recognize a specific 8-mer PBS.
List of vectors containing or expressing ORFs of Casilio modules
Plug-and-play functional genomics
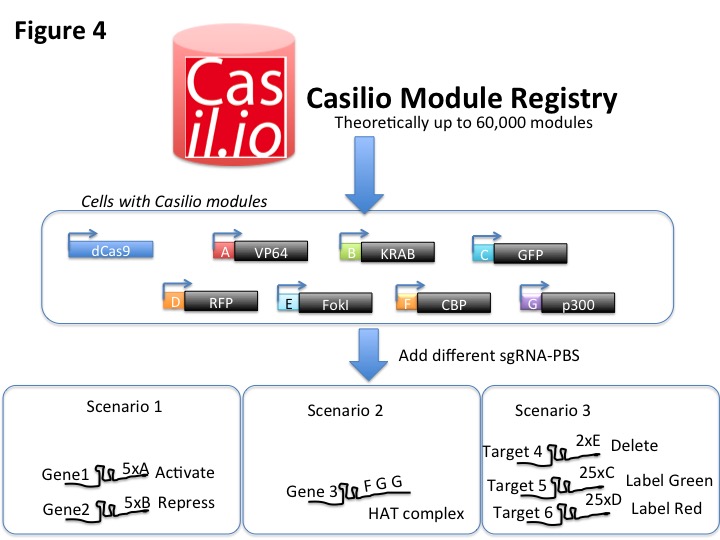
Due to the programmable feature of PUF/PBS, Casilio has the potential to allow 60,000 orthogonal modules to function independently in cells. Modules tethering different fusion proteins will be assigned a unique PUF/PBS pairing. We will maintain a Casilio module reigstry with links to resources and information. Researchers can obtain the desired modules to perform their functional genomic experiments by supplying the matching sgRNA-PBS. Since Casilio modules are orthogonal, researchers can use any combinations of modules as is, allowing plug-and-play for the application of Casilio in functional genomics studies (Fig 4).
Future development
To make Casilio more useful, we will construct lentiviral vectors for expressing Casilio components and generate orthogonal Casilio modules with different effector functions.
References
1 . Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816-821.
2 . Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife 2013; 2:e00471.
3 . Mali P, Yang L, Esvelt KM et al. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-826.
4 . Wang H, Yang H, Shivalila CS et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153:910-918.
5 . Cong L, Ran FA, Cox D et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819-823.
6 . Qi LS, Larson MH, Gilbert LA et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:1173-1183.
7 . Cheng AW, Wang H, Yang H et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 2013; 23:1163-1171.
8 . Hilton IB, D’Ippolito AM, Vockley CM et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015.
9 . Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013; 10:977-979.
10 . Gilbert LA, Larson MH, Morsut L et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154:442-451.
11 . Perez-Pinera P, Kocak DD, Vockley CM et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 2013; 10:973-976.
12 . Chen B, Gilbert LA, Cimini BA et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479-1491.
13 . Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 2013; 10:1116-1121.
14 . Mali P, Aach J, Stranges PB et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013; 31:833-838.
15 . Konermann S, Brigham MD, Trevino AE et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2014; 517:583-588.
16 . Zalatan JG, Lee ME, Almeida R et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015; 160:339-350.
17 . Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 2015.
18 . Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014; 159:635-646.
19 . Cheng AW, Jillette N, Lee P et al. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res 2016.
20 . Chen Y, Varani G. Engineering RNA‐binding proteins for biology. FEBS J 2013; 280:3734-3754.
21 . Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nature communications 2012; 3:1147.
22 . Wang Y, Cheong C-G, Hall TMT, Wang Z. Engineering splicing factors with designed specificities. Nat Methods 2009; 6:825-830.
23 . Wang Y, Wang Z, Tanaka Hall TM. Engineered proteins with Pumilio/fem‐3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J 2013; 280:3755-3767.